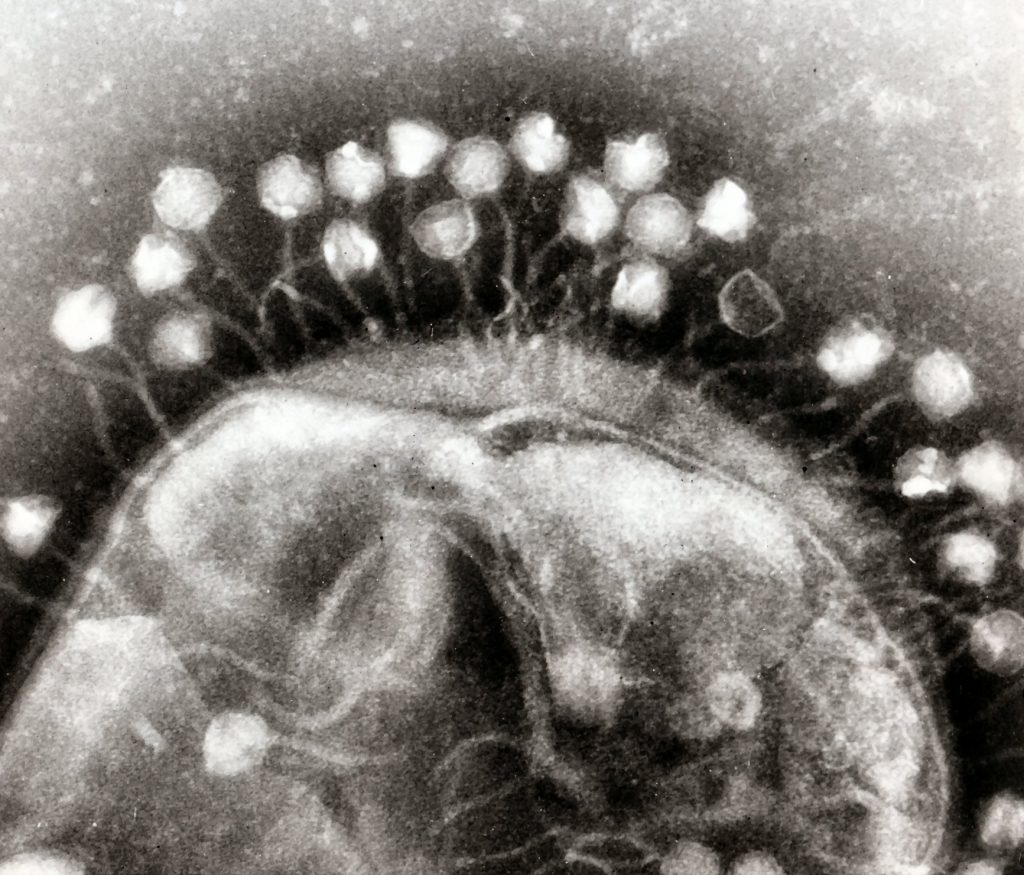
Bacteriophage cluster around a bacterium, ready to hijack its resources. Image: Dr Graham Beards|Wikimedia.
Dr Adam Middleton, a researcher at Otago Biochemistry, introduces bacteriophage viruses and explains how he uses them to help with his research into molecules involved in cancer.
The bacteriophage – ‘the deadliest being on earth’ – is a virus that infects bacteria and forces the bugs to dedicate the remainder of their brief lives to generate more phage. Eventually the bacteria burst and out emerge heaps of phage.
Phage consist of nucleic acid (DNA or RNA) contained in a protein shell. They attach to the outside of bacteria, and inject their guts into the cell. Because they are highly specific for their target bacteria – and do not infect humans – phage have been used in desperate attempts to save the lives of people infected with antibiotic-resistant bacteria. In Eastern Europe they are commonly used to treat bacterial infections.
Why are we talking about phage?
I recently spent three months at the University of Toronto working in the laboratory of Professor Sachdev Sidhu where I learned how to perform a technique called ‘phage display’.
Coincidentally, half of the 2018 Nobel Prize in Chemistry was given to two scientists for their work with phage display: George P. Smith for developing the technique, and Sir Gregory P. Winter for using phage to develop antibodies.
What exactly is phage display?
Phage display is a way to make new proteins. Proteins are large complex molecules that your cells make to build, fix, and maintain your body. For example, haemoglobin is a protein that transports oxygen from your lungs to your cells.
Scientists can use phage display to help them make new proteins, which can be used to treat diseases such as cancer. However, instead of designing every detail of a protein, phage display lets you ‘harness the power of evolution’ using bacteria and phage.
The best example is for antibodies. When bacteria or viruses infect you, your body uses antibodies to target the invasion. Part of this is a process of ‘hypermutation’ of antibodies, where your immune cells mutate parts of the antibodies to develop ones that are highly specific to the target.
What scientists can do (and this is why Sir Winter got the Nobel prize), is to mutate antibodies in a test tube (and end up with billions of variants), then put these into phage. The phage are then used as platforms to display the antibody, and used for selection against your target protein of interest.
To do this selection, you take advantage of the simplicity of phage. Phage are more-or-less packages of DNA or RNA surrounded by a protein coat.
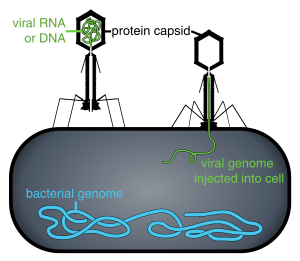
Phage injecting its genome into bacteria. Image: Adenosine|Thomas Splettstoesser|Wikimedia.
Scientists can make a fusion between a coat protein and their protein of interest (eg: an antibody) by putting the DNA encoding their protein in line with the DNA for the coat protein. This DNA is transcribed and translated, and results in a chimeric protein-coat protein. This fusion is then excreted from the phage to form the coat, and results in your protein being ‘displayed’ on the phage.
The next step is to infect bacteria with the phage, and allow the bacteria to replicate. The result is billions of bacteria and billions of phage. The phage is removed from the bacteria, and used for selection against your desired target protein(s).
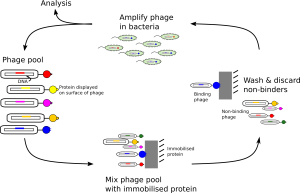
How to select the phages that bind best to the target protein. If you repeat the cycle several times, the resulting phages should bind very tightly to their target proteins. Image: Adam Middleton.
Selection goes something like this: 1) immobilise your protein of interest; 2) mix your phage with your immobilised protein; 3) wash away non-binding phage; 4) remove bound phage; 5) infect bacteria and allow them to replicate; 6) remove phage from bacteria; and 7) repeat steps 2-6 three or four times. The result is phage that bind tightly to your target protein, and each phage contains the DNA that encodes the tightly-binding protein. Congratulations! You have made and isolated a new protein that binds tightly to your target. You can now investigate its biological effect!
What can you use phage display to do?
What I did in Toronto was use phage display to investigate the small molecule ubiquitin.
Ubiquitin is a very important protein that underlies most activities of the eukaryotic cell. It modifies substrate proteins in a highly-controlled manner to direct their fate. Some kinds of ubiquitin modifications direct substrates to be part of large protein complexes (bunches of protein), some are involved in regulating gene transcription, and some direct the substrate protein to be degraded.
My goal is to trap the formation of this degradation signal by mutating ubiquitin and use phage display to find variants that bind to other protein components of the ubiquitin machinery.
What emerged from Toronto is a new library containing billions of variants that can be displayed on phage. These will help us understand how degradative ubiquitin chains can be formed, and how this process can be manipulated.
Disruptions of ubiquitin transfer are associated with many diseases – such as cancer – and targeting protein degradation is an established therapeutic technique. By understanding more precisely how to control the ubiquitin code, we hope to be able to develop more specific ways of targeting the ubiquitin system.

