 300-level biochemistry student Hannah Gordon writes about the potential uses of cells that we design ourselves and the spontaneously organising molecules that we must understand before we can make artificial cells.
300-level biochemistry student Hannah Gordon writes about the potential uses of cells that we design ourselves and the spontaneously organising molecules that we must understand before we can make artificial cells.
Can you imagine a self-assembling, low maintenance system that converts household waste into fuel to heat and power your home? A system that could also be applied to the production of lifesaving pharmaceuticals? Converting one chemical to another is called metabolism, mastered by the molecular machines of life – cells. Could their clever strategies be copied and altered to create designer cells that produce compounds of our choosing?
In this feature, we will explore promising applications for synthetic cells and the difficulty in their design. Building a synthetic cell from the ground up requires a comprehensive understanding of the processes which fulfil the requirements for life, particularly a process called ‘self-organisation’. To appreciate the complexity of these processes, we will walk through one of the simplest and best studied self-organising systems – regulation of cell division by Min proteins.
Why should we synthesise cells?
Cells that can be made artificially have great potential in many areas, particularly in medicine and in solving environmental issues.
Increasingly, we use living molecules to treat disease. Insulin, the hormone that helps us regulate sugar absorption, is a classic example of a treatment best produced by living machinery. People with diabetes either do not make insulin themselves or cannot use it efficiently, so sometimes need to be given insulin to control their disease. Extracting and purifying insulin from pigs meant diabetics could live with this once-fatal disease. But pig insulin is not quite the same as human insulin, and purification was not always perfect, sometimes causing an allergic reaction.
We can now genetically engineer bacteria cells to produce human insulin, which reduces the contamination risk and production cost. However, a cell’s metabolism is highly complex, and altering it to produce high quantities of something it has not evolved to make may lead to harmful by-products and high mortality rates of the cells.
Now, with an advanced understanding of molecular biology, scientists are looking to create a synthetic template cell from which living machines could be designed to produce a variety of compounds such as insulin. Built with only components essential to life, in theory, the problems encountered when a complex metabolism is tampered with could be dodged.
Though this seems far-fetched, very simple cell-like structures have been synthesised in the lab. Protocells, which are self-organised spherical membranes thought to be precursors to the first living cells, are involved in trials for delivering cancer drugs to the site of tumours.
The prospects for synthetic cells are not limited to the pharmaceutical industry. Attempts to alter the metabolism of various single-celled organisms to produce biofuels have been met with similar issues to insulin synthesis. The possibility of designer cells has inspired concepts of buildings with “organs” – a compartment of synthetic cells designed to convert waste (such as carbon dioxide, greywater etc.) into heat and power for a building. This could be hidden or made a feature.

Microalgae façades that generate renewable energy from algal biomass and solar thermal heat, designed by construction-engineering cooperative Arup.
Architects have also proposed the design of synthetic cells to form reefs. Cells designed to move away from light and produce “biocrete” added to the waterways of Venice could strengthen the city’s foundations and slow its sinking.
What makes a cell living?
To engineer a living cell, first one must ask, “What is life?” Schrödinger wrote that life is the creation of order in a world that tends towards disorder. Living things avoid decay into chaos by exchanging energy with their environment.
Consider your body as a factory. Its energy consumption allows you to do work but is also necessary for the management of all its machines. Your cells function autonomously, and the inner workings of these miniature machines – proteins – self-organise into complex patterns that avoid a natural tendency towards disorder.
Self-organisation occurs when separate, disordered parts become ordered by interacting with each other. It happens when there is enough energy available, but there is no control from anything outside those parts. For example, people in a stadium making a Mexican wave.
The ability for the molecules inside cells to self-organise is essential for life. We need to understand how they do this so we can create cells from scratch.
Some proteins that self-organise so cells divide: the Min system
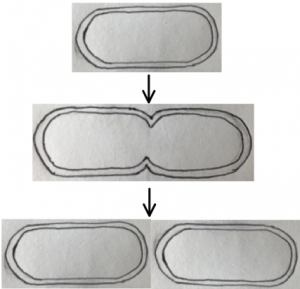
Consider one essential requirement for life: cell reproduction. To reproduce, a cell must divide evenly into two identical daughter cells, and this division has to be as efficient as possible. For companies designing miniature pharmaceutical factories, efficient cell division ensures maximum turnover. In bacteria, cell division is regulated by the highly studied Min system. MinC, D and E are proteins that self-organise together, making sure that their cell will divide evenly at its middle. This relatively simple system could be useful to include in a synthetic cell. It may also inform design of other self-organising systems. Let’s explore this simple example of self-organisation to appreciate the intricacy of the inner workings of cells, the machines of life.
What’s the name of the game?
Understanding a biochemical system is like learning to play a video game. Any good gamer will have an idea of the virtual world and how characters can move within it. They will then explore possible interactions between characters, working towards the aim or end goal of a storyline. Consider the E. coli bacterial cell: a three-dimensional, rod-shaped world. The environment inside the cell is mostly water-like, and it is surrounded not by layers of atmosphere but by a membrane.
Proteins are the tiny characters that inhabit this world and mostly move freely through this environment, though some can attach to the membrane. We’ll meet some key characters in a bit.
First, what is the aim of this storyline? The cell can divide to produce two new cells. For these to be functional, the division must occur at the middle of the original cell.
Our first character is a protein called FtsZ. There are many copies of FtsZ in a cell that arrange as a team into a structure that pulls the cell apart. FtsZ has the power to divide the world into two, and left unchecked it could cause its demise. The three protagonists, the Min proteins, restrict FtsZ to where it is useful.
Introducing MinC, the gifted child
The protein MinC is the only character that can directly interact with FtsZ. Perhaps you could think of FtsZ as wild creatures that only MinC have the power to tame. MinC binds FtsZ in a way that renders it unable to bind other FtsZ proteins.
But MinC’s power needs almost as much regulation as FtsZ. In our storyline, it would be a gifted child whose ability to tame creatures should be controlled. After all, with no wild FtsZ, how would the world become two? MinD is the guardian of MinC. They move together around the cell, much like a parent giving a child a piggyback.
Introducing MinD, the guardian
MinD, the guardian of MinC, keeps it away from the middle of the cell by moving from one end to the other. Throughout the timeline, MinD transverses the long axis of the world so that come time for division, MinC is mostly at the poles, and FtsZ is wild in the middle.
None of our characters can move in a controlled way through the water-like environment but float about randomly. To move along the cell, MinD can cling to the membrane, jump off, and then cling on again. The lag in their ability to grasp the bordering membrane again means they have to float some distance before doing so. To understand how this occurs in a directed manner, let’s meet MinD’s partner, MinE.
Introducing MinE, the shy lawyer
To grasp to the membrane, two copies of MinD must work as a pair. You could imagine one giving the other a leg up. MinDs are not particularly trusting, and require a temporary contract to form an alliance. This “contract” is a little molecule called adenosine triphosphate or ATP. MinE is the external party required for the MinDs to “break” their contract.
It takes some time to redraft a contract, hence there is a lag before reattachment. MinD and MinE are the ultimate team. Not only does MinE initiate separation and detachment of MinDs, but membrane-bound MinD activates MinE. You could imagine MinE as a shy lawyer, holding its pen to its chest. Approaching MinD, it is emboldened, flinging its arms wide ready to reach out and sign that release waiver. Once confident, MinE lingers to initiate detachment of more MinDs.
A Mexican Wave!
Now that we understand the roles of each character, let’s think about the overall gameplay again. There are many copies of each type of Min protein in the cell, and membrane attachment and detachment of these proteins results in a pattern of movement like a standing wave.
Consider a reverse Mexican wave: imagine a stadium full of people who have their arms up (representing MinD proteins attached to the membrane). Someone representing MinE moves along each row to coordinate the timing of people bringing their arms down. Coordinators hang back to ensure all hands are down before moving on to the next column. People put their arms back up on their own, but with a delay. When the coordinators reach the end of the stand, they turn around and the wave continues the other way. The geometry of the bacterial cell is such that, on average, most of the MinDs are attached to the ends of the cell. MinC’s ability to stop FtsZ is enhanced when MinD is bound to the membrane, thus cell division is restricted to the middle of the cell.

A membrane targeting sequence rescues pattern formation by slowing MinE detachment from MinD. Image adapted from Surovtsev & Jacobs-Wagner (2018) Subcellular Organization: A Critical Feature of Bacterial Cell Replication https://doi.org/10.1016/j.cell.2018.01.014
A toolkit for building self-organising proteins
For biochemists and gamers alike, understanding the rules and interactions of players is never enough. A deeper understanding of the game can be gained through analysis of a player’s character. What characteristics are indispensable?
Recent research showed that MinE and MinD proteins mixed together interact to create a lovely spiralling pattern (a beautiful example of self-organising):
 But when the researchers made a shortened MinE that consisted of only the part it needs to bind to MinD, then mixed it with MinD, no pattern was seen:
But when the researchers made a shortened MinE that consisted of only the part it needs to bind to MinD, then mixed it with MinD, no pattern was seen:
 Previous research suggested that MinE can stick to other MinE proteins (dimerisation) and also bind the membrane. So, the researchers tried adding back the parts of MinE that enable it to dimerise or stick to the membrane. This allowed it to make a variety of patterns when mixed with MinD again:
Previous research suggested that MinE can stick to other MinE proteins (dimerisation) and also bind the membrane. So, the researchers tried adding back the parts of MinE that enable it to dimerise or stick to the membrane. This allowed it to make a variety of patterns when mixed with MinD again:


 Images adapted from Glock et al. Design of biochemical pattern forming systems from minimal motifs. eLife 2019;8:e48646. https://doi.org/10.7554/eLife.48646
Images adapted from Glock et al. Design of biochemical pattern forming systems from minimal motifs. eLife 2019;8:e48646. https://doi.org/10.7554/eLife.48646
Using mathematical modelling to confirm, they concluded that the different parts of MinE favour different mechanisms for pattern formation. Adding a dimerisation motif to the minimal MinE rescues pattern formation by increasing its recruitment to MinD. Adding a membrane-targeting sequence rescues pattern formation by slowing MinE detachment from MinD.
This research gives new purpose to a well-studied protein system. By identifying the contribution of different protein modules to pattern formation, a toolkit of modules could be created. Proteins could be designed to organise into specific patterns. For designing a synthetic cell for home organs, this would be valuable for gathering together proteins necessary for the biofuel metabolic pathway. When proteins in a reaction pathway are close, the reaction is faster. The self-organising modules could be temperature sensitive so that when the building is cold, the proteins cluster and more biofuel is produced.
Not only does Min show promise for inclusion in a template synthetic cell, but it could also inform the design of other self-organising protein systems.
But exploring the potential to engineer life is fraught with ethical concerns. Should we be playing God? Both risks and benefits should be carefully weighed. As with genetically modified organisms, there are concerns around what could happen if synthetic cells are not appropriately contained. Would production of biofuels that significantly reduce our environmental impact be worth this risk? Would more efficient pharmaceutical production make life-saving drugs more accessible or simply line the pockets of the rich?
We are still some way off being able to design synthetic cells capable of these things. In the meantime, scientists continue to explore systems such as Min simply to better understand the incredible processes of life.
Further reading:
Designing with Protocells: Applications of a Novel Technical Platform
https://www.mdpi.com/2075-1729/4/3/457
Subcellular Organization: A Critical Feature of Bacterial Cell Replication
https://doi.org/10.1016/j.cell.2018.01.014
Design of biochemical pattern forming systems from minimal motifs
https://doi.org/10.7554/eLife.48646
How biologists are creating life-like cells from scratch
https://www.nature.com/articles/d41586-018-07289-x