 An article written by 300-level Biochemistry student Emily Crosse about research by PhD student Jordon Lima.
An article written by 300-level Biochemistry student Emily Crosse about research by PhD student Jordon Lima.
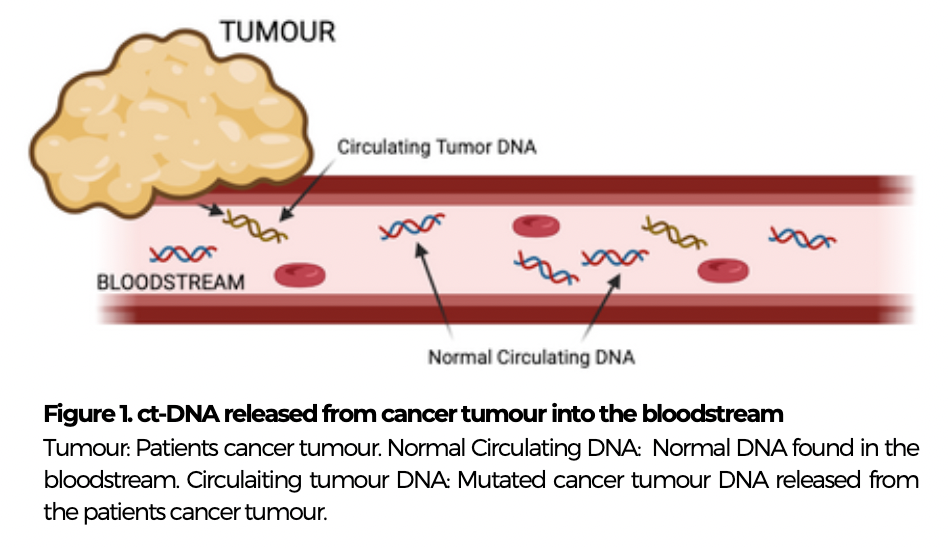
Circulating tumour DNA (ct-DNA), is cell-free DNA released from tumours into the bloodstream, with a short half-life, making it a good potential marker for easy detection of colorectal cancer in patients, via the bloodstream. Currently tissue biopsies are performed to identify cancer, however this is a difficult and invasive detection method, contributing to the lack of early diagnosis of colorectal cancer and challenges in therapeutic monitoring.
Along with this, CT and MRI scans are used for detection and monitoring of cancer through the quantification of changes in metabolic activity and variations in tumour size. These current detection methods have limitations due to their cost and accessibility, creating barriers to cancer treatments and diagnostics for people within the community. This contributes to the frightening colorectal cancer statistics in New Zealand.
Liquid biopsy of ct-DNA may be able to provide an early detection and intervention method for colorectal cancer patients, as well as allowing the long term monitoring of therapeutic responses, in a non-invasive manner. This is because ct-DNA can provide a global snapshot of a patients cancer, directly from a blood sample. Knowing this, Jordon Lima and her research team at Otago University have begun a research investigation, with the aim to convince people that ct-DNA needs to be added to routine surveillance and detection of cancer. In particular, the focus for Lima is to help reduce barriers to cancer treatment and diagnosis for people in Māori communities, as they are more at risk of fatality due to colorectal cancer.
The paper Zou et al. (2020) provides the basis for the investigation into ct-DNA’s potential for surveillance, as opposed to an alternative biological marker, carcinoembryonic antigen (CEA). CEA is a blood protein marker which is currently used for cancer detection, however is limited by its specificity and sensitivity. This paper provides valuable insight and justification for research into diagnostic capabilities of ct-DNA, due to its sensitivity and specificity. This was identified through ct- DNA analyses over a clinically relevant timeline of cancer patients, identifying that ct-DNA provided a more sensitive and responsive measurement of cancer tumours, with 97% of patients having detectable ct-DNA, in comparison to 83% patients returning positive detection of CEA. Additionally, ct-DNA was identified as being cancer tumor specific, and therefore provides a more detailed method of detection and monitoring.
Meet the Scientist
Jordon Lima, an Otago University Biochemistry PhD student, has set off on her investigation into ct-DNA and its potential for diagnostic use in cancer care, with her project Māu tēnā kīwai o te kete, māku tēnei.
The initiation of this project started in her postgraduate honours year, looking at the mutations in RNF43, and her discovery of the link between this gene and colorectal cancer. Commonly colorectal cancer arises from a mutation in the APC gene, accounting for around 80% patients, however little was known about the other 20% of patients. Therefore, Lima’s breakthrough discovery of the link between RNF43 and colorectal cancer was an exciting discovery, helping scientists to understand more about the other 20% of colorectal cancer patients.
With colorectal cancer being the second deadliest cancer, due to the lack of early diagnosis and difficulty in tracking the disease, Lima’s investigation into RNF43 and the link to this disease lead to the young scientist wanting to further discover how these statistics can be improved for the New Zealand population. More specifically, reducing the barriers to cancer care for the Māori community. Māori is highly representative of these statistics, due to their barriers faced in cancer care, including access, language and diagnosis. The project ‘Māu tēnā kīwai o te kete, māku tēnei’ means to each share a handle of the basket. In this case, one handle is held by the scientists and researchers behind the study of ct-DNA sensitivity and specificity, and its potential for use in cancer care. The other handle represents Lima’s communication and discussion with people in Māori communities, discovering their barriers to cancer treatment and diagnostic. Therefore, the basket connecting these handles displays the barriers to cancer care faced by Māori communities.
 Nanopore Sequencing
Nanopore Sequencing
Jordon Lima and her team are investigating how ct-DNA could be used to reduce the barriers to treatment and diagnosis of colorectal cancer for people in Māori communities through the use of Nanopore sequencing. Nanopore sequencing is a third-generation sequencing method, which functions by feeding a DNA sample through a pore from one side of a membrane to another, which interrupts an electrical current, creating peaks and troughs in the current. These peaks and troughs correspond to different DNA bases passing through the pore, allowing a minimum DNA sequencing accuracy of 99.6% (https://nanoporetech.com/accuracy). These nanowire sequencers are pocket size and relatively low cost, allowing them to be easily taken out into the community for DNA sequencing.
This provides exciting capabilities for use in detection of colorectal cancer through being able to measure ct-DNA by looking at the mutations detected in the DNA sequencing. The hope for Lima and her team is to utilise this new technology to detect these mutations and quantify ct-DNA levels in patients out in the community in real time. Therefore being able to improve the early diagnosis of colorectal cancer and allow easy therapeutic monitoring.
The future of this science and technology could provide the capability to reduce these barriers for people in the Māori community in a cost-effective, highly sensitive and efficient manner. Routine usage of this technology and ct-DNA as a blood sample biomarker for cancer detection is a scientific discovery which should not be overlooked!
References:
Lima, J. (2022). Equitable Application of ct-DNA to the NZ Population O. U. T. Presentation. Otago University.
Oliveira, K. C. S., et al. (2020). “Current Perspectives on Circulating Tumor DNA, Precision Medicine, and Personalized Clinical Management of Cancer.” Mol Cancer Res 18(4): 517-528.
Zou, D., et al. (2020). “Circulating tumor DNA is a sensitive marker for routine monitoring of treatment response in advanced colorectal cancer.” Carcinogenesis 41(11): 1507-1517.


