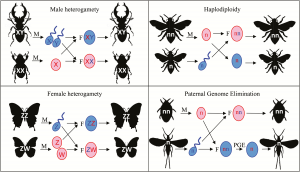
The determination of sex within many animal species is determined by specific genetic differences, with a set of un-matched chromosomes. Of these chromosomes one is smaller or completely absent whereas the other contains a full complement of genes. In mammals these chromosomes are the X chromosome and the smaller Y chromosome. As this is the only clear genetic difference, it is likely that all sex differences originate from this imbalance or the epigenetic impact of these chromosomes.
Understanding how these chromosomal differences related to the sex differences we see is a question of particular interest as they are the key cause of sex differences and lead to many major changes. As such, this information can be used to help inform any research based around this field. While the focus of this is primarily on the XY sex chromosome pair, it is worth being aware other types exist.
Evolving the Sex Chromosomes
The evolution of this XY chromosome pair in mammals, dates back to roughly 300 million years, originating within a pair of autosomes (non-sex chromosomes).[2] In mammals this lead to the XY pair found in males, whereas bird show ZW pairing in females which has evolved separately. This is believed to be caused by the emergence of SRY, the male determining gene, on the future Y chromosome. The evolution of this region was the beginning of turning this autosome pair into sex chromosomes.
Following on from this, the proto-Y chromosome would gain a series of inversions where regions in the chromosome a reversed. The prevents proper chromosomal crossing over during meiosis and allowed for the full divergence of the X and Y chromosomes that has occurred since. Pairing only occurs thanks to a para-autosomal region (PAR) at the ends of both the X and Y chromosome, keeping the chromosomes together as a pair and can even have crossing over within this region.[3]
Without proper chromosomal segregation, the Y chromosome develops male specific regions which do not match with the X chromosome. Alongside this the Y chromosome would suffer genetic decay and losing 97% of ancestral genes in humans.[4]
How Genes Create Sex Differences
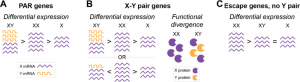
One of the key differences between the two different chromosomes is the genes found on them. Because of the variable dosage sensitivity on these genes and the process of X Chromosome Inactivation (XCI), in which one of the two X chromosomes in females are inactivated, there is variety of different types of genes on the sex chromosomes.
Five types of sex chromosome genes include
PAR1 Genes
PAR1 Genes are found in the shared para-autosomal region on both the X and Y chromosome.[5] While these genes are not the source of sex differences, unique phenotypes related to sex chromosomes such as Turner’s are likely to have major impacts from these genes as the maintenance of genes within this region implies a strong dosage sensitivity.
X-Y Pair Genes
X-Y pair genes have the next strongest dosage sensitivity, with a high level of dosage sensitivity maintaining expression of these genes on both the X and Y chromosomes.[6] Due to the fact these genes are not maintained through crossing over however, over time there is variation that develops within these genes between the X gene and Y gene. The evolutionary divergence within these genes can lead to one being more or less efficient than the other or gaining additional roles.[7,8]
These evolutionary differences at the gene level can translate across into sex differences due to the impacts this can have on various pathways.
X-Inactivated GenesX-Inactivated genes had a middling level of dosage sensitivity. While still required, these genes could be lost off the Y chromosome during evolution. To keep the level of these genes consistent across both sexes, one X chromosome is inactivated randomly in cells with multiple X chromosomes.[10] While this generally keeps levels consistent, there is a point before a X chromosome has been inactivated in which this difference in dosage can have an impact. This unequal level of gene expression can have dramatic impacts on the product depending on the downstream effects it might have.[11]
Escape Genes
Escape genes have the lowest dosage sensitivity. As such they are still lost on the Y chromosome. However, due to this low dosage these genes are able to possibly escape X-inactivation. This in turn allows these genes to have unequal between males and females, allowing for them to contribute to sex differences.[12]
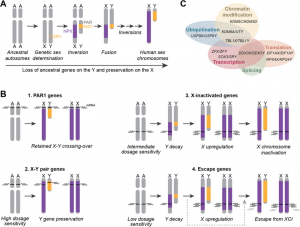
Diagram displaying evolution of both XY chromosomes (A) and the origins of the different types of gens found on them (B). Function roles of X-Y paired genes are also displayed in Venn Diagram (C)[9]
Imprinted genes have a unique effect on sex differences. In males, the X chromosome is inherited maternally and as such they lack any paternal X imprinting. When X inactivation occurs, it only inactivates one copy of the X chromosome, but this is not consistent across cells. As such, females gain a patchwork of maternal and paternal imprinting, whereas males will only inherit maternal imprinting. This means males will have more maternal imprinting than females, while paternal imprinting will only appear in males.[13] Autosomal Genes
Autosomal genes can also impact sex differences. This is in response to the different levels of sex chromosome genes found within the body, which can act as a type of on-off switch for autosomal genes. Through the activating of these genes, a many different functions can be activated, allowing for complexity in variation. All these differences in expression however must trace back to the sex chromosomes and either the genetic or hormonal changes induced.[14]
Researching Sex Chromosome Variation
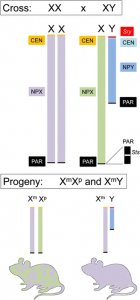
To study these genes, the main method use is by looking at what are known as the “Four Core Genotype” mice.[15] In these mice, the SRY region is moved from the Y chromosome onto one of the autosomes. This means that the hormonal changes associated with SRY are no longer associated with XY chromosome. This creates a set of four different types of mouse, with mice being either XX female, XX male, XY female, or XY male.[13]
Looking at the phenotypes of these four different mice can then be used to separate whether phenotypes are hormonally induce (and thus only show up in mice with the SRY gene), direct effects (stemming from X or Y chromosome) or affected by both. By identifying these effects, the origins of these differences can be discovered by looking into genes that could affect these pathways.[13]
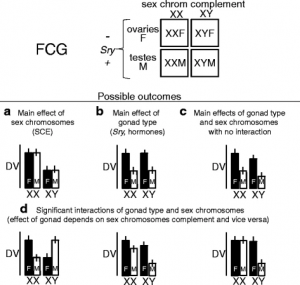
This diagram displays the different results we can get on phenotypic effects from a four core genotypes mouse cross. This information can help inform if sex chromosomes have an impact and potentially the type of gene.[13]
Studies of this in beetles with XY chromosomes, found that the evolution of sexual dimorphism was quicker when the selection pressure was on male beetles rather than females. The mechanism through which this occurs is not currently known, however two hypotheses presented in the study say that it is either due to dominance of some genes being associated with whether the sex of the individual or that recessive traits on the X chromosome could be selected for in males, whereas females select for the dominant trait. This would lead to selection for heterozygous females and males with the recessive X being selected for.[16]
An Example of Gene Impacts
One gene identified as have an effect on sex differences is the short stature homeobox (SHOX) gene. This genes is responsible for the growth of longer bones in human limbs and the lack of this gene in patients with Turner’s Syndrome (who only have a single X chromosome and no Y) is one of the most well characterised effects of the disease.[17] Located in PAR, loss of this gene off the arm of X chromosome was found to be the cause of short stature and an extra copy being found in those with taller stature.[18,19] There is evidence suggesting that there is a difference in SHOX between males and females, alongside evidence suggesting that even a small difference in expression could still have a non-zero impact on height.[20]
Conclusion
Overall evidence on the evolution of sex chromosomes has the chromosomes evolving from an autosomal pair following the gain of a male specific region in mammals. Following this, matching of the two sex chromosomes was lost and variation between these chromosomes was gained over time. Various types of genes can be found on these chromosomes, with dosage sensitivity being an important factors in preventing decay on the shorter chromosome. Separation of direct chromosomal effects from effects stemming from hormonal differences is critical to building our understanding of the role of sex chromosomes in the evolution and development of sex differences.
For more information on sex differences, other pages covering the evolution of sex differences and the impacts of these differences on different organs such as the brain and heart can be found on the left side of the screen.
References
[1] Blackmon, H., Ross, L., & Bachtrog, D. (2017). Sex determination, sex chromosomes, and karyotype evolution in insects. Journal of Heredity, 108(1), 78-93.
[2] Lahn, B. T., & Page, D. C. (1999). Four evolutionary strata on the human X chromosome. Science, 286(5441), 964–967. https://doi.org/10.1126/science.286.5441.964
[3] Freije, D., Helms, C., Watson, M. S., & Donis-Keller, H. (1992). Identification of a second pseudoautosomal region near the Xq and Yq telomeres. Science, 258(5089), 1784–1787. https://doi.org/10.1126/science.1465614
[4] Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., Chinwalla, A., Delehaunty, A., Delehaunty, K., Du, H., Fewell, G., Fulton, L., Fulton, R., Graves, T., Hou, S. F., Latrielle, P., … Page, D. C. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature, 423(6942), 825–837. https://doi.org/10.1038/nature01722
[5] Ciccodicola, A., D’Esposito, M., Esposito, T., Gianfrancesco, F., Migliaccio, C., Miano, M. G., Matarazzo, M. R., Vacca, M., Franzè, A., Cuccurese, M., Cocchia, M., Curci, A., Terracciano, A., Torino, A., Cocchia, S., Mercadante, G., Pannone, E., Archidiacono, N., Rocchi, M., Schlessinger, D., … D’Urso, M. (2000). Differentially regulated and evolved genes in the fully sequenced Xq/Yq pseudoautosomal region. Human molecular genetics, 9(3), 395–401. https://doi.org/10.1093/hmg/9.3.395
[6] Bellott, D. W., Skaletsky, H., Cho, T. J., Brown, L., Locke, D., Chen, N., Galkina, S., Pyntikova, T., Koutseva, N., Graves, T., Kremitzki, C., Warren, W. C., Clark, A. G., Gaginskaya, E., Wilson, R. K., & Page, D. C. (2017). Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nature genetics, 49(3), 387–394. https://doi.org/10.1038/ng.3778
[7] Lahn, B. T., & Page, D. C. (1997). Functional coherence of the human Y chromosome. Science, 278(5338), 675–680. https://doi.org/10.1126/science.278.5338.675
[8] Skaletsky, H., Kuroda-Kawaguchi, T., Minx, P. J., Cordum, H. S., Hillier, L., Brown, L. G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., Chinwalla, A., Delehaunty, A., Delehaunty, K., Du, H., Fewell, G., Fulton, L., Fulton, R., Graves, T., Hou, S. F., Latrielle, P., … Page, D. C. (2003). The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature, 423(6942), 825–837. https://doi.org/10.1038/nature01722
[9] San Roman, A. K., & Page, D. C. (2019). A strategic research alliance: Turner syndrome and sex differences. American journal of medical genetics. Part C, Seminars in medical genetics, 181(1), 59–67.
[10] Jegalian, K., & Page, D. C. (1998). A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature, 394(6695), 776–780. https://doi.org/10.1038/29522
[11] Richardson, V., Engel, N., & Kulathinal, R. J. (2023). Comparative developmental genomics of sex-biased gene expression in early embryogenesis across mammals. Biology of sex differences, 14(1), 30. https://doi.org/10.1186/s13293-023-00520-z
[12] Tukiainen, T., Villani, A. C., Yen, A., Rivas, M. A., Marshall, J. L., Satija, R., Aguirre, M., Gauthier, L., Fleharty, M., Kirby, A., Cummings, B. B., Castel, S. E., Karczewski, K. J., Aguet, F., Byrnes, A., GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, … MacArthur, D. G. (2017). Landscape of X chromosome inactivation across human tissues. Nature, 550(7675), 244–248. https://doi.org/10.1038/nature24265
[13] Burgoyne, P. S., & Arnold, A. P. (2016). A primer on the use of mouse models for identifying direct sex chromosome effects that cause sex differences in non-gonadal tissues. Biology of sex differences, 7, 68.
[14] Petropoulos, S., Edsgärd, D., Reinius, B., Deng, Q., Panula, S. P., Codeluppi, S., Plaza Reyes, A., Linnarsson, S., Sandberg, R., & Lanner, F. (2016). Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell, 165(4), 1012–1026. https://doi.org/10.1016/j.cell.2016.03.023
[15] De Vries, G. J., Rissman, E. F., Simerly, R. B., Yang, L. Y., Scordalakes, E. M., Auger, C. J., Swain, A., Lovell-Badge, R., Burgoyne, P. S., & Arnold, A. P. (2002). A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. The Journal of neuroscience : the official journal of the Society for Neuroscience, 22(20), 9005–9014. https://doi.org/10.1523/JNEUROSCI.22-20-09005.2002
[16] Kaufmann, P., Wolak, M. E., Husby, A., & Immonen, E. (2021). Rapid evolution of sexual size dimorphism facilitated by Y-linked genetic variance. Nature ecology & evolution, 5(10), 1394–1402.
[17] Clement-Jones, M., Schiller, S., Rao, E., Blaschke, R. J., Zuniga, A., Zeller, R., Robson, S. C., Binder, G., Glass, I., Strachan, T., Lindsay, S., & Rappold, G. A. (2000). The short stature homeobox gene SHOX is involved in skeletal abnormalities in Turner syndrome. Human molecular genetics, 9(5), 695–702. https://doi.org/10.1093/hmg/9.5.695
[18] Rao, E., Weiss, B., Fukami, M., Rump, A., Niesler, B., Mertz, A., Muroya, K., Binder, G., Kirsch, S., Winkelmann, M., Nordsiek, G., Heinrich, U., Breuning, M. H., Ranke, M. B., Rosenthal, A., Ogata, T., & Rappold, G. A. (1997). Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nature genetics, 16(1), 54–63. https://doi.org/10.1038/ng0597-54
[19] Ogata, T., & Matsuo, N. (1993). Sex chromosome aberrations and stature: deduction of the principal factors involved in the determination of adult height. Human genetics, 91(6), 551–562. https://doi.org/10.1007/BF00205079
[20] Tukiainen, T., Villani, A. C., Yen, A., Rivas, M. A., Marshall, J. L., Satija, R., Aguirre, M., Gauthier, L., Fleharty, M., Kirby, A., Cummings, B. B., Castel, S. E., Karczewski, K. J., Aguet, F., Byrnes, A., GTEx Consortium, Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group, Statistical Methods groups—Analysis Working Group, Enhancing GTEx (eGTEx) groups, NIH Common Fund, … MacArthur, D. G. (2017). Landscape of X chromosome inactivation across human tissues. Nature, 550(7675), 244–248. https://doi.org/10.1038/nature24265