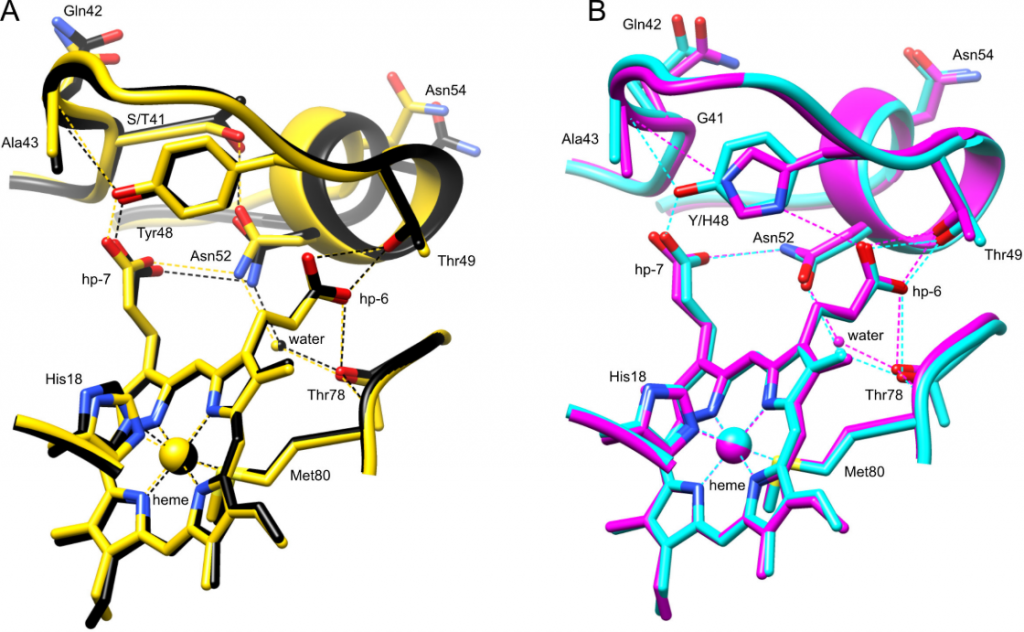
Cytochrome c variants
H-bond networks in cytochrome c variants. Comparison of the hydrogen bonding network between the human cytochrome c 40-57 Ω loop (residues 39-57 shown) and the heme for (A) G41T (black, chain A, PDB ID 6ECJ) and G41S (yellow, chain B, 3NWV), and (B) WT (cyan, chain A, 3ZCF (13)) and Y48H (magenta, chain A, 5EXQ).
Supervisor: Dr. Fellner (Biochemistry) & Dr. Ledgerwood (Biochemistry)
The cytochrome c project is in collaboration with the Ledgerwood Lab. Co-supervision between the Fellner and Ledgerwood laboratory at the Department of Biochemistry will strengthen this project.
Summary: This project is ideal if you want to use protein crystallography and other structural tools like molecular dynamics to study the structural changes based on disease causing mutations at the atomic scale. Linking the biochemical studies of human cytochrome c variants in the Ledgerwood lab with in vitro characterisation of purified cytochrome c in the Fellner lab will form a strong thesis project for any interested student. Several cytochrome c variants have succesfully been produced and some crystallised in the Fellner laboratory, including using obtained structures for molecular dynamics. Here is where you come in as several variants have not been characterised in this way, making for an exciting thesis project. This project also includes measurements at the Melbourne Australian synchrotron (in person or remotely depending on your preference).
Further reading:
Fellner, M., Parakra, R., McDonald, K. O., Kass, I., Jameson, G. N. L., Wilbanks, S. M., Ledgerwood, E. C. (2021) Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity, Biochem Journal, 478, 669-684. DOI: 10.1042/BCJ20200793