This page summarises our research on Pseudomonas aeruginosa universal stress proteins (USPs), please see the individual pages for details about available student projects (summer, undergraduate, PhD). If you are interested please email matthias.fellner@otago.ac.nz.
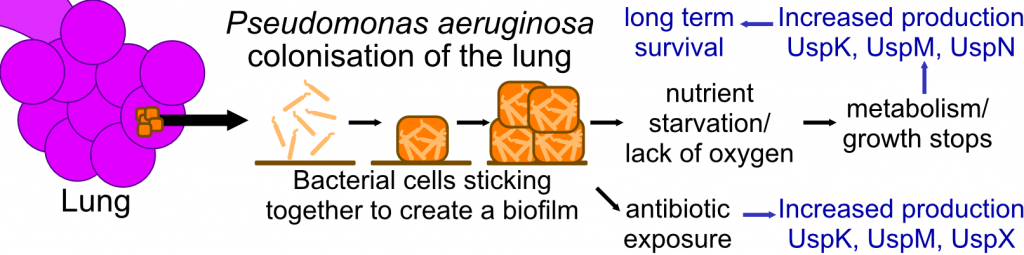
The steady and significant increase in serious Pseudomonas aeruginosa infections in New Zealand, coupled with notable sociodemographic disparity in disease incidence, is a disturbing national trend. Studies into the underlying protein biochemical processes are critical to improve this situation in the long-term. P. aeruginosa is an opportunistic bacterium that has the ability to inhabit various environmental niches. It can adapt to many stress conditions that would kill other bacteria during infections. This stress adaptation, especially encountered by the bacterium during chronic infections in patients, has been linked to specific proteins, called universal stress proteins (USPs). We aim to establish the first initial characterisation of these proteins in vitro. This will enable future drug development strategies targeting universal stress proteins to cure P. aeruginosa infections in patients, which account for one third of the annual deaths caused by bacterial pathogens in New Zealand.
P. aeruginosa biofilm formation in lungs, survival prolonged by upregulation of USPs. Adopted from (1)
USPs have been detected in many organisms including plants, archaea, metazoans, and bacteria.(1) They function in a series of various cellular responses to stressful conditions such as oxidative stress, exposure to DNA damaging agents, nutrient starvation, high temperature and acidic stress.(1) Although a highly conserved group of proteins, the molecular and biochemical aspects of their exact functions are largely unknown, however it has been shown that USPs are essential for P. aeruginosa to persist during chronic infections.
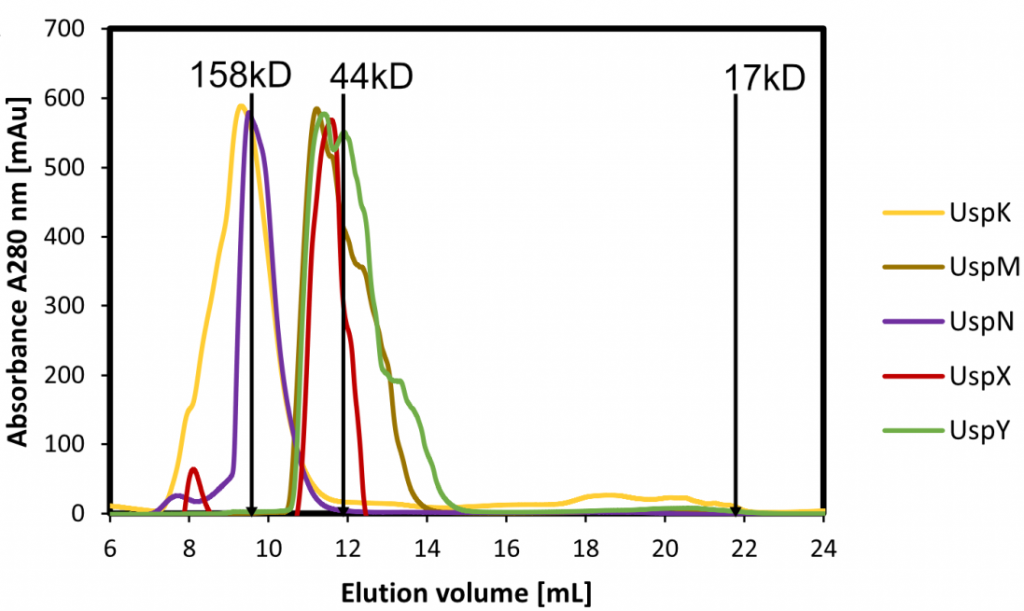
We successfully produced five P. aeruginosa USPs to a high yield and purity for in vitro studies. The USP domain is a highly conserved 140–160 amino acid-containing domain that may exist in P. aeruginosa as a single ~15kD (UspK, UspX, UspY) or as a tandem repeat ~30kD (UspM, UspN). Compared to standard proteins of 158kD and 44kD size it is apparent that all P. aeruginosa USPs form oligomers, UspK ~dodecamer, UspM ~dimer, UspN ~pentamer, UspX ~tetramer and UspY ~tetramer.
Size exclusion chromatography of P. aeruginosa USPs, elution compared to standard proteins indicates oligomeric forms.
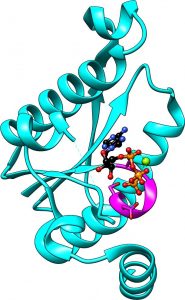
USPs are also closely related to the PP-loop ATP pyrophosphatase family, showing an identical secondary structure fold but lacking the conserved PP-loop residues for catalysis. The PP-loop binds ATP and uses it for substrate adenylylation, with the substrate normally being bound by an adjacent substrate binding domain. We have given a structural perspective on the 18 enzyme family members containing this PP-loop (2) following characterisation of one of the most unique members, a sacrificial sulfur transferase, including structures of bound ATP explaining the biological function and reaction mechanism of this enzyme.
USP related domain from sacrificial sulfur transferase bound by ATP-Mg to the PP-loop in magenta (PDB ID 5UDS) (3)
We hypothesise that USPs also bind nucleotides and presented them to other proteins for catalysis. This protein-protein interaction would mimic how the PP-loop ATP pyrophosphatase family functions via internal domain-domain interactions. The resulting catalysis would be responsible for the observed essential function of USP proteins and give us a better understanding what to target in future work in drug development.
(1) Masamba, P.; Kappo, A. P., Parasite Survival and Disease Persistence in Cystic Fibrosis, Schistosomiasis and Pathogenic Bacterial Diseases: A Role for Universal Stress Proteins? Int J Mol Sci 2021, 22. DOI: 10.3390/ijms221910878.
(2) Fellner, M.; Hausinger, R. P.; Hu, J., A structural perspective on the PP-loop ATP pyrophosphatase family. Crit Rev Biochem Mol Biol 2018, 53, 607-622. DOI: 10.1080/10409238.2018.1516728.
(3) Fellner, M.; Desguin, B.; Hausinger, R. P.; Hu, J., Structural insights into the catalytic mechanism of a sacrificial sulfur insertase of the N-type ATP pyrophosphatase family, LarE. Proc Natl Acad Sci U S A 2017, 114, 9074-9079. DOI: 10.1073/pnas.1704967114.