This page summarises our research on Staphylococcus aureus serine hydrolases, please see the individual pages for details about available student projects (summer, undergraduate, PhD). If you are interested please email matthias.fellner@otago.ac.nz.
Staphylococcus aureus is a common cause of a variety of diseases ranging from local skin or soft tissue infections to invasive and chronic infections such as bacteremia, pneumonia, or endocarditis. For 2019 S. aureus was the leading bacterial cause of death in 135 countries (including New Zealand), being the only organism that was associated with more than 1 million deaths worldwide.(1)
There is an urgent need for better diagnostic tools such as advanced imaging agents to determine where exactly the infection is located, how significant it is and if it is responding to antibiotic treatments.
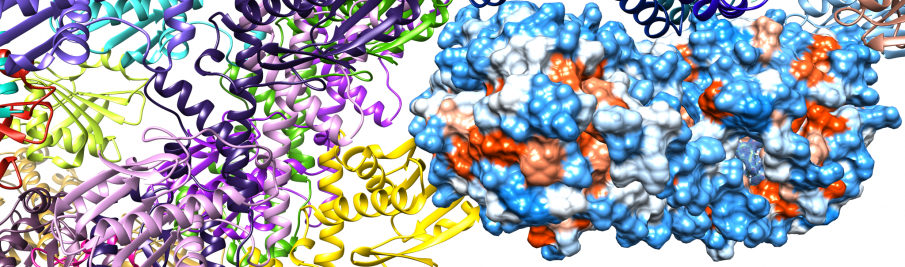
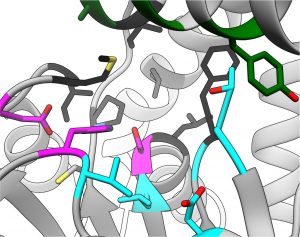
We create such tools by covalently targeting ten S. aureus serine hydrolase proteins (FphA-J 52-22 kD). These proteins are active during S. aureus infections, making them promising targets for diagnostics.(2) Our strategy ensures that the new diagnostic tool will have exceptional levels of specificity, negating off-target toxic effects while also offering additional therapeutic benefits due to the inhibition of the target proteins.
Fluorophosphonate-binding hydrolases A-J
- All active in S. aureus biofilms
- Cut unknown molecules
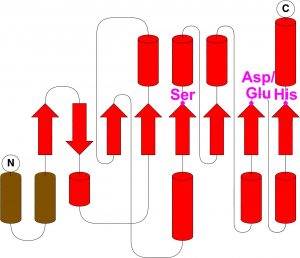
- Cutting by activated serine (S-H-D/E)
Share common fold, except N-terminal helices and first N-terminal β strand
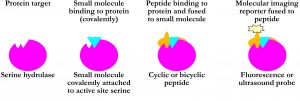
General strategy:
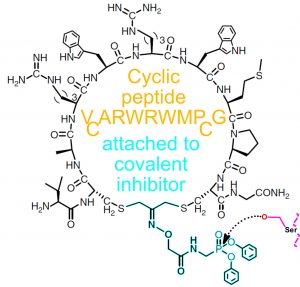
Utilisation of the serine hydrolase catalytic mechanism: Stalling the enzyme in a covalent intermediate state by introducing certain small molecules. Specificity improved by attachment of small molecules to target specific cyclic or bicyclis peptides.
A very brief summary about progress on each protein:
For available student projects on individual proteins please see dedicated pages.
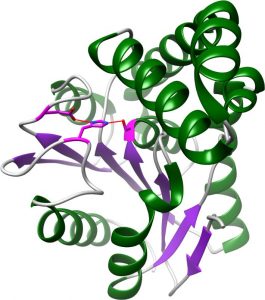
FphA:
- FphA is a lot bigger than other members (α-helices a lot longer)
- Appears to have very specific substrate preference
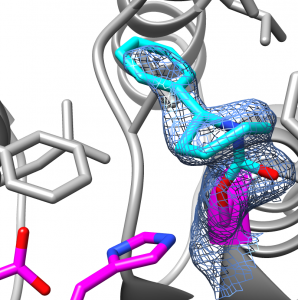
- Successfully produced and crystallised in the Fellner laboratory
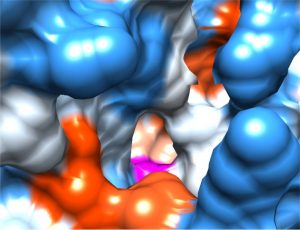
Unique FphA active site, Ser in magenta
FphB:
- FphB contains N-terminal helices, most likely for membrane binding
- Truncated versions successfully produced in the Fellner laboratory
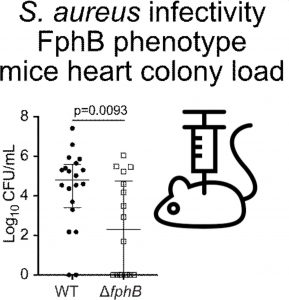
Mice clear infection faster when fphB is disrupted
Chloroisocoumarin inhibitor with fluorescent suggest membrane binding
FphC
- FphC highly conserved in staphylococci
- Lower abundance than other Fph proteins
- Difficult to target and to produce recombinantly
FphD
- FphD contains N-terminal helices, in contrast to FphB most likely to fold onto the active site
- Truncated versions successfully produced in the Fellner laboratory
FphE
- FphE least conserved (only in ~50% of staphylococci)
- Ideal target to differentiate between pathogenic (ex S. aureus) and non-pathogenic strains (ex S. epidermidis)
- A wide range of small molecules that specificially bind to FphE have been disocovered
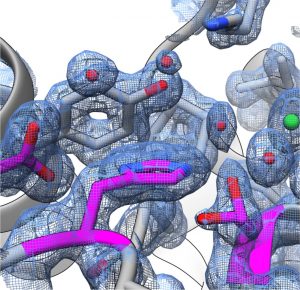
- Successfully produced and crystallised in the Fellner laboratory at high resolution in complex with many ligands
Example: Boronic acid based molecule bound FphE structure 1.6Å
FphF
- FphF might be the most abundant Fph protein during S. aureus life cycle
- A highly effective cyclic peptide based inhibitor has been identified for FphF
- A wide range of small molecules that specificially bind to FphF have been disocovered
- Successfully produced and crystallised in the Fellner laboratory at high resolution in complex with many ligands
Example: Triazole urea based molecule bound FphF structure 1.9Å
FphG
- FphG highly conserved in staphylococci
- Lower abundance than other Fph proteins
- Difficult to target and to produce recombinantly
FphH
- Most conserved Fph, even across all Bacillales
- Correlation with stress response of S. aureus
- Invovled in antibiotic defense – have discovered cleavage of antibiotic fusidic acid
- A wide range of small molecules that specificially bind to FphH have been disocovered
- Successfully produced and crystallised in the Fellner laboratory at high resolution
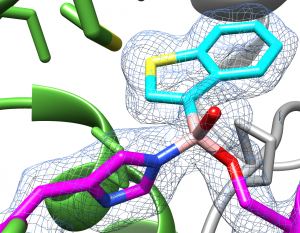
FphH active site at 1.6Å
FphI
- Unique staphylococci distant homolog of FphH (28% identity, closest match among Fph proteins)
- Slower enzyme kinetics compared to FphH suggests preference for a unique different substrate
- Successfully produced and crystallised in the Fellner laboratory at ultra-high resolution
FphH active site at 1.1Å
FphJ
- FphJ is significantly smaller than other Fph proteins
- Very short α-helices suggest that active site is very different to other Fph proteins
- Successfully produced in the Fellner laboratory
Two additional secreted S. aureus serine hydrolases SAL1 + SAL2 have been discovered decades before the Fph proteins, these are also being studied in the Fellner laboratory, both have successfully been produced and SAL2 has been crystallised.
(1) Collaborators, G. B. D. A. R., Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 2023, 400, 2221-2248. DOI: 10.1016/S0140-6736(22)02185-7
(2) Lentz, C. S.; Sheldon, J. R.; Crawford, L. A.; Cooper, R.; Garland, M.; Amieva, M. R.; Weerapana, E.; Skaar, E. P.; Bogyo, M., Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP). Nat Chem Biol 2018, 14, 609-617. DOI: 10.1038/s41589-018-0060-1
Additional reading about Fluorophosphonate-binding hydrolases A-J (52-22 kD):
Fellner, M., Walsh, A.; Dela Ahator, S.; Aftab, N.; Sutherland, B.; Tan, E. W.; Bakker, A. T.; Martin, N. I.; van der Stelt, M.; Lentz, C. S. (2023) Biochemical and Cellular Characterization of the Function of Fluorophosphonate-Binding Hydrolase H (FphH) in Staphylococcus aureus Support a Role in Bacterial Stress Response. ACS Infectious Diseases (in print). DOI: 10.1021/acsinfecdis.3c00246
Fellner, M., Lentz, C. S., Jamieson, S. A., Brewster, J. L., Chen, L., Bogyo, M., Mace, P. D. (2020), Structural Basis for the Inhibitor and Substrate Specificity of the Unique Fph Serine Hydrolases of Staphylococcus aureus. ACS Infectious Diseases, 6, 2771-2782. (Chosen as the cover article) DOI: 10.1021/acsinfecdis.0c00503
Fellner, M. (2021) Newly discovered Staphylococcus aureus serine hydrolase probe and drug targets. ADMET & DMPK, 10, 107-114. DOI: 10.5599/admet.1137
Keller, L. J.; Lentz, C. S.; Chen, Y. E.; Metivier, R. J.; Weerapana, E.; Fischbach, M. A.; Bogyo, M., Characterization of Serine Hydrolases Across Clinical Isolates of Commensal Skin Bacteria Staphylococcus epidermidis Using Activity-Based Protein Profiling. ACS Infect Dis 2020, 6, 930-938. DOI: 10.1021/acsinfecdis.0c00095
Additional reading about Fph small molecule and peptide discovery:
Chen, L.; Keller, L. J.; Cordasco, E.; Bogyo, M.; Lentz, C. S., Fluorescent triazole urea activity-based probes for the single-cell phenotypic characterization of Staphylococcus aureus. Angew Chem Int Ed Engl 2019, 58, 5643-5647. DOI: 10.1002/anie.201900511
Chen, S., Lovell, S. D., Lee, S., Fellner, M., Mace, P. D. Bogyo, M. (2021), Identification of highly selective covalent inhibitors by phage display. Nature Biotechnology, 39, 490-498. DOI: 10.1038/s41587-020-0733-7