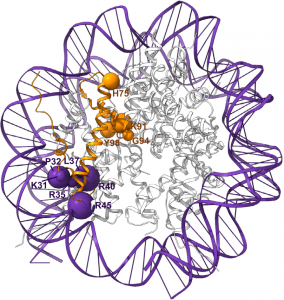
A model of histone H4 disease-causing amino acid changes (orange ribbon) either cluster to the N-terminal α-helix facing toward DNA (cluster 1, purple spheres) or are located in regions buried within the nucleosome core (cluster 2, orange spheres). Size of sphere indicates the relative prevalence of substitutions affecting that residue.
Supervisor: Dr. Fellner (Biochemistry) & Dr. Bicknell (Biochemistry)
The histone H4 project is in collaboration with the Bicknell Lab. Co-supervision between the Fellner and Bicknell laboratory at the Department of Biochemistry will strengthen this project.
Summary: This project is ideal if you want to use protein crystallography to study the structural changes based on disease causing mutations at the atomic scale. Linking the study and discoveries of histone H4 variants in the Bicknell lab with in vitro characterisation of histones in the Fellner lab will form a strong thesis project for any interested student. H3-H4 dimers (including H4 variants), histone octamers and nucleosomes have succesfully been produced by Dr. Fellner in the past. Building on these preliminary results the next step will be to obtain protein crystals. Characterisation of the purified proteins using other biochemical techniques, as well as genetic studies in the Bicknell lab will also be option. This project also includes measurements at the Melbourne Australian synchrotron (in person or remotely depending on your preference).
Further reading:
Tessadori, F., Duran, K., Knapp, K., Fellner, M.,.. Bicknell, L. (and 60 others) (2022) Recurrent de novomissense variants across multiple histone H4 genes underlie a neurodevelopmental syndrome, American Journal of Human Genetics, 109, 750-758. DOI: 10.1016/j.ajhg.2022.02.003