Wayne Patrick, a senior lecturer in the Otago Department of Biochemistry*, is a devoted fan of evolution and proteins. He and his former PhD student, Matilda Newton, along with colleagues in Sweden, have been looking at changes that happen as an organism evolves, not just in the genes, but also in the machinery of cells, the proteins.
Thanks to Darwin and others we know a great deal about evolution, but there is still a lot to learn, particularly the nitty-gritty at the molecular level. We need to understand how evolution works, not just to understand how people and other creatures came into existence, but for many reasons, especially to improve our understanding and treatment of diseases.
Patrick and his colleagues used Salmonella bacteria to study evolution happening in real time. Yes, Salmonella are the bacteria that give people horrible food poisoning. The scientists who set up the original experiment several years ago worked with a strain of the nasty bacteria under control in the lab.
Like all living things, bacteria are constantly evolving, only at a pace where we can watch the action happening, so they make a good model to study evolution in action.
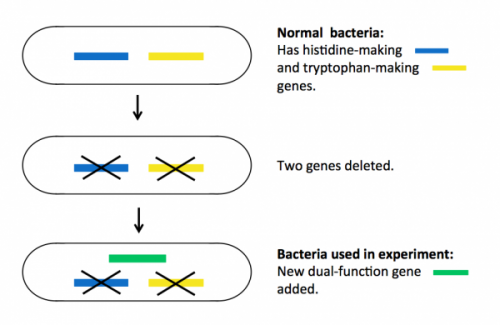
Bacteria (as well as all living things) need the amino acids histidine and tryptophan to grow and multiply. Normally, they have proteins inside them that make these amino acids, so if you take histidine and tryptophan away from their food, they will survive happily.
To prepare for the experiment, the researchers deleted two genes in the bacteria, each encoding a protein essential for making either histidine or tryptophan. These mutant bacteria die if histidine and tryptophan are taken away from their food.
Next they put a single new ‘dual-function’ gene back into the bacteria. This gene encodes a mutant protein that is able to do the jobs of both of the two deleted proteins, but very inefficiently. The new bacteria can survive without histidine and tryptophan in food, but only just – they do not thrive.
Here’s how the researchers set up the rest of their experiment:
- They fed the new bacteria food that didn’t contain any histidine and tryptophan, forcing the bacteria to make their own histidine and tryptophan using the inefficient dual-function gene. They then let the bacteria grow and multiply for thousands of generations.
- Over the course of about 3000 generations, the researchers took regular samples of the evolving bacteria, and sequenced their DNA to see what changes happened to the dual-function gene.
- Patrick and his colleagues took these gene sequences, and made the proteins that they encode. They then tested how well the different proteins worked and looked at the exact shape and structure of each protein.
So what changes did they see as the bacteria evolved? They monitored changes in three areas:
- how well the bacteria survived with their depleted food source
- what happened to the dual-function gene
- what happened to the protein encoded by the dual-function gene
At first, the weak dual-function gene let the bacteria survive, but not very well – the bacteria took as long as five hours to double their population. However, after several hundred generations the bacteria started doing a lot better. The doubling time dropped dramatically to about two hours, showing that the bacteria were evolving to thrive even though no histidine and tryptophan was provided.
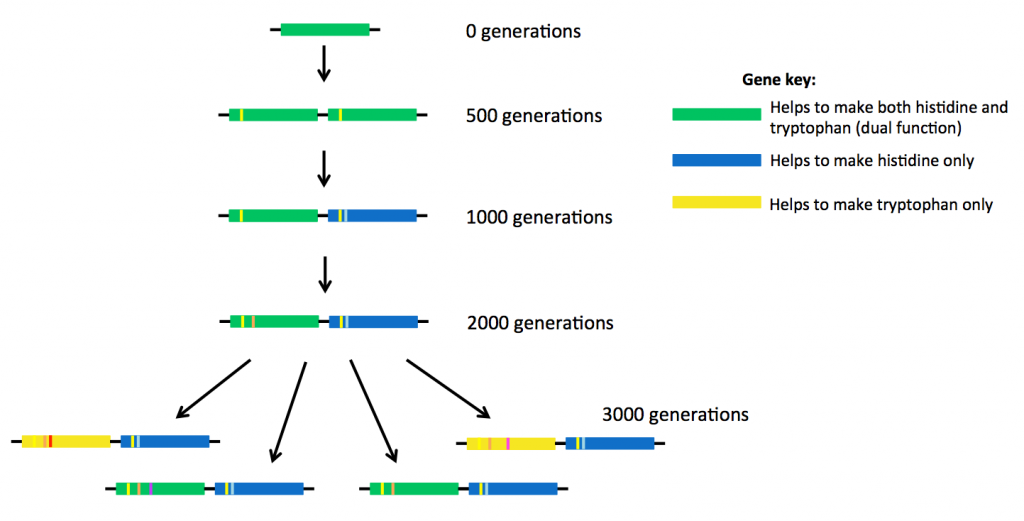
At the same time, the researchers also saw the ‘birth’ of two new genes.
After 500 generations, the bacteria had made a copy of the original weak dual-function gene, so that there were now two identical genes present, sitting side-by-side in the DNA (see above). After about 1000 generations, small mutations were starting to accumulate in the two genes, different mutations in each gene.
Of course, changes in the genes meant that changes were also occurring in the proteins made from the genes.
The original ancestor protein made from the dual-function gene (the top gene coloured green in the picture above) was able to carry out two jobs. But the proteins descended from that ancestor started to have slightly different shapes and therefore different abilities to do these jobs.
Eventually some of these new proteins became specialised. Some of them continued to carry out two reactions (encoded by green genes above), but some proteins could only help to make histidine (blue genes). Other proteins specialised in making tryptophan (yellow genes). The new proteins were much more efficient at doing their jobs than their ancestor.
What were the changes in the shapes of the proteins that caused them to perform differently?
The original ancestor protein looked like a blob, as many proteins do, with a backbone in the shape of what’s known as a ‘TIM barrel’:
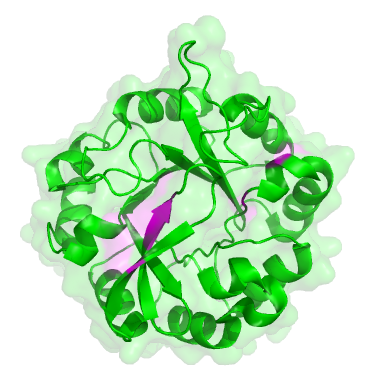
The original dual-function protein from the start of the experiment. Places where mutations occured are marked in pink.
To the untrained eye, all of the descendent proteins looked the same as the original. But they had subtle changes, in the parts coloured pink in the picture of the green protein here. These changes were mainly in the active site, the part of the protein that carries out the chemical reaction, i.e. the ‘business’ end of the protein. The rest of the protein is structural, there to hold the active site parts in the right place.
These small changes resulted in big differences in the way the proteins carried out their chemical reactions, and therefore how well the bacteria survived and multiplied. The researchers could directly relate the changes in the shape of the proteins to the improved growth of the bacteria as they evolved.
The researchers also found that the changes that evolved in the proteins were not ones that could have been guessed or calculated beforehand by people. Evolution is still far ahead of human technology when creating innovative new protein micro-machines.
Through their intricate work, Patrick and his colleagues have helped to capture snapshots of a protein evolving in real time. They have revealed previously unseen details of evolution at the molecular level and demonstrated what a complicated and unpredictable business it can be.
Read the full paper here:
Structural and functional innovations in the real-time evolution of new (βα)8 barrel enzymes. Matilda S. Newton, Xiaohu Guo, Annika Söderholm, Joakim Näsvall, Patrik Lundström, Dan I. Andersson, Maria Selmer, and Wayne M. Patrick. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(18):4727-4732. DOI: 10.1073/pnas.1618552114
See also:
Real-Time Evolution of New Genes by Innovation, Amplification, and Divergence. Joakim Näsvall, Lei Sun, John R. Roth and Dan I. Andersson. Science. 2012; 338 (6105): 384-387. DOI: 10.1126/science.1226521
*Wayne Patrick is now an Associate Professor at the Victoria University of Wellington.