Genes within the DNA of humans, plants & yeasts are actually interrupted with bits of non-gene DNA scattered throughout, and these bits are called introns. Otago Biochemistry’s Associate Professor Chris Brown and Dr Chun Shen Lim delve into the evolution of an intriguing genetic phenomenon…
Since the surprising discovery over 40 years ago that genes from complex organisms, e.g. humans, plants, yeasts, and mushrooms, were split into pieces by introns, there has been ongoing debate as to why. Indeed, the pieces of most genes need to be spliced back together to work. This seemed a very inefficient system — raising questions about why such a near-universal system evolved, and what its advantages might be.
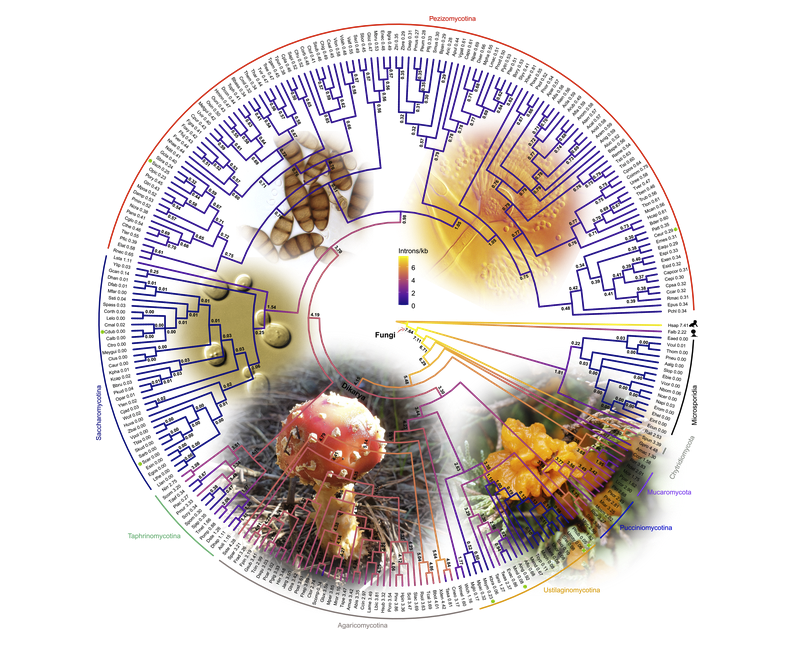
Strikingly, the 19,000 protein-coding genes in our genomes have over 150,000 of these intervening introns that need to be spliced out. Most of these introns have no known functions. Why are we so inefficient? In contrast, the humble bakers’ or brewer’s yeast has few introns, less than 300, but most of yeast’s distant relatives, notably mushrooms, have tens of thousands. It seems that the ancestor of all fungi, including yeasts, disease-causing smuts, and mushrooms, might have had many introns and that these have been lost during evolution, as proposed by the ‘introns early’ hypothesis.
To address this enigma, an international team led by Dr Chun Shen Lim and Assoc Prof Chris Brown at the University of Otago with Brooke Weinstein and Dr Scott Roy from the University of California-Merced, used hundreds of genomes from diverse fungi to reconstruct the evolutionary histories of these introns. This was made possible by advanced genomic analysis approaches. The research article is available as a preprint and will be published on 10th Oct 2021 in Molecular Biology and Evolution from the Oxford University Press.
These fungi include beneficial organisms (e.g. bakers yeast), edible mushrooms, and also fungi that cause disease (e.g. the smuts and rusts of crops) or human pathogens (causing >1 million deaths/year)
The new results supported the idea that the ancient common ancestor of fungi (about 900 million years ago) was intron-rich, supporting the once hotly debated idea of ‘introns first’ or ‘introns early’. Surprisingly, the new study also estimated more introns in the fungal ancestor than previous studies. There were many more episodes of intron loss over intron gain through the millennia.
Dr Lim and the team then went on to ask, what is special about the few introns that have been retained in most organisms? This analysis was enabled by the finding that a number of fungi have lost most of their introns. By looking for introns that are still in those, the team found that those introns with essential functions are retained. Some of the most interesting examples are introns as genes themselves, which is analogous to Russian dolls, or ‘genes within genes’, that are related to core biological functions.
Intriguing and as yet unanswered questions are — what do the few introns that are kept do? Do they function in a similar way, or in some new way? Can these functions be used as antifungal targets in plant fungal diseases? These questions may be able to be addressed by experimental testing in yeast or by even deeper comparative genomics. Ongoing projects by Assoc Prof Brown and colleagues are using a new generation of sequencing technology and genomic approaches to address this and other fundamental biological and evolutionary questions.
In related studies, Dr Lim has just (23/9/2021) been awarded a University of Otago Research Grant for 2022 ( $36,147) with Associate Investigator Brown. The study is entitled “Deciphering translation of the ‘untranslated’ regions of messenger RNAs”, where they will investigate the roles of introns in more complex organisms — humans and plants.
Chun Shen Lim, Brooke N Weinstein, Scott W Roy, Chris M Brown. Analysis of fungal genomes reveals commonalities of intron gain or loss and functions in intron-poor species. Molecular Biology and Evolution, 10 Oct 2021. https://doi.org/10.1093/molbev/msab094